代表性成果 (获奖成果、专著、论文、专利) |
代表性成果: (1)从一级序列到三维结构对蛋白质进行数学表征,如物化性质、构象、能量等方面进行解析,结合特征评估与筛选算法,利用支持向量机、随机森林等机器学习方法进行模型构建。 分析功能位点的各种序列、结构及进化信息,完成了对不同蛋白质功能位点的数字表征及模型构建(Comput Bio Chem, 2012, 4, 304-312; Comput Bio Chem, 2012, 42, 1053-1059);采取差异性分析思路对蛋白质功能进行注释(Chemometr Intell Lab 2012,110, 163-167; PLoS ONE, 2013, 8, e84439; Chemometr Intell Lab, 2013, 126, 117-122;Analyst, 2015, 140, 3048-3056;RSC adv, 2017, 31, 2046-2069);基于碱基组成差异分析预测miRNA的dSNP (Interdiscip Sci: Comput life Sci, 2017, 9, 459-467)。 提出了蛋白质互作表征新方法,如基于domain-domain相互作用的预测模型(Chemometr Intell Lab, 2014, 136, 97-103; J Comput Aid Mol Des, 2014, 44, 699-703); 引入信息论方法,将信息熵应用到蛋白质功能位点的保守性表达分析(Sci Rep, 2015, 5, 12403; J Comput Aid Mol Des, 2017, 31, 1029-1038)。 利用变构效应和能量变化参数,结合晶体结构分析,对蛋白质相互作用面进行分类研究,并实现其作用强弱的定量预测(Proteins, 2014, 82, 3090-3100; J Comput Aid Mol Des, 2014, 28, 619-629;Chemometr Intell Lab, 2014, 138, 7-13; Sci Rep, 2015, 5, 14214 );对活性口袋进行结构解析,对其可药性进行预测(Curr Pharm Design, 2015, 21, 3051-3061)。 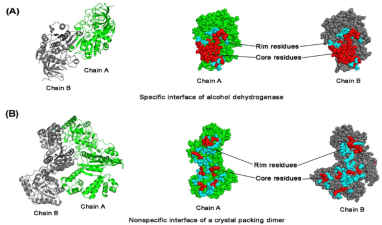
提出Pbase(RNA 碱基埋藏面积占 RNA 总结合面积的比例)参数将不同的蛋白质-RNA复合物聚为三类(RSC Adv, 2018, 8, 10582-10592);提出一种基于弹性距离阈值来定义RNA结合残基的新方法,称为两倍最小距离法2dmin (J Comput Aid Mol Des, 2018, 32, 1363-1373)。该方法考虑复合物之间的个体差异来确定不同复合物的距离阈值,并以此利用随机森林算法构建RNA结合位点预测模型,其预测准确率达90%。 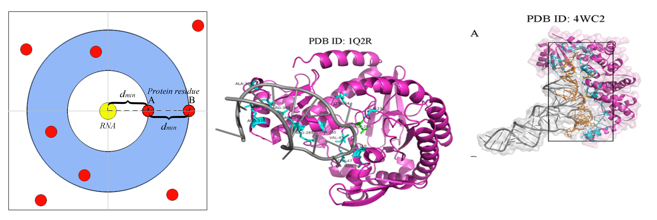
(2)利用分子网络方法,对癌症关联miRNA的作用机制进行研究。 解析17种癌症相关miRNA在其靶标蛋白与互作蛋白相互作用网络调控中的作用(Sci Rep, 2016, 2016, 6, 34172)。网络分析表明,越多miRNA调控的靶标蛋白在功能上的差异性小,相反其相似性就越高。对卡波氏肉瘤相关疱疹(KSHV)病毒编码的miRNA靶标的功能做了细致的分析(Sci Rep, 2017, 7, 3159)。16个miRNA调控的蛋白共表达网络可划分为六个重要的功能模块,均被证明与KSHV的发病机制高度相关。 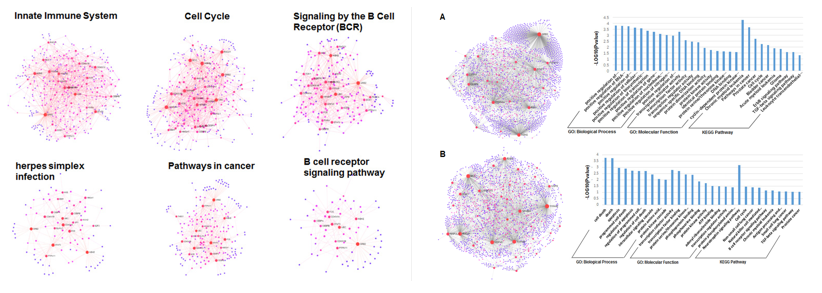
利用分子网络技术,实施基于miRNA的肾癌分子分型研究(Brief Bioinform, 2020, 1, 73-84 )。构建miRNA关联的ceRNA-ceRNA 相互作用网络,分析三种肾癌亚型(KICH, KIRC和 KIRP)之间的差异图谱,筛选hub基因,考察它们在不同亚型中的作用机制。利用网络计算结合基因表达实验确认SETD2、TRIP11与VHL可作为对KIRC的分型标记。调控网络分析给出三个基因之间的相互调控关系。 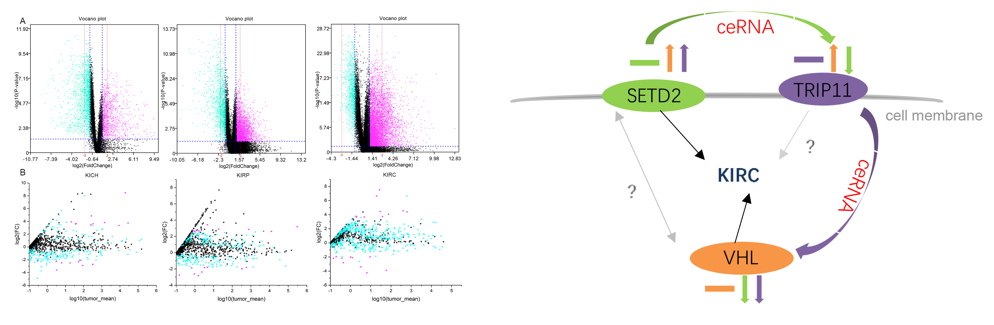
开发癌症血液外泌体的ceRNA网络数据库。针对三种癌症,包括结直肠癌(CRC),肝细胞癌(HCC)与胰腺癌(PAAD),利用超几何测试、网络拓扑分析及hub节点功能分析,获取百万级ceRNA相互作用对。数据库名为ExoceRNA Atlas (//www.exocerna-atlas.com/exoceRNA)。网站FB体育如下图所示。 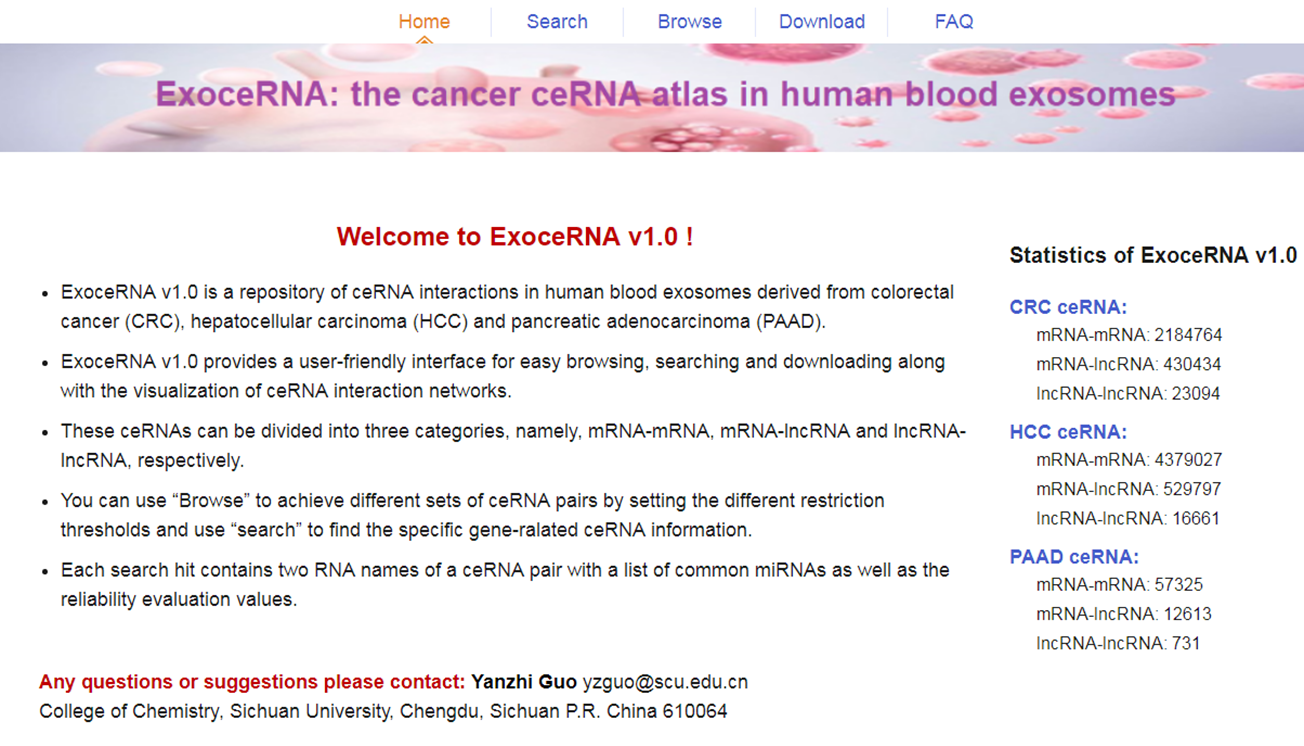
(3)利用生物多组学数据分析提取分子特征,结合病人临床数据与机器学习算法实现对不同癌症的风险预测。 DNA基因甲基化位点的变化能调控基因的表达,通过分析差异性甲基化位点与差异性基因mRNA、lncRNA及miRNA之间的关系,计算筛选7个DNA甲基化位点,并利用LASSO-Cox算法实现对KIRC肾癌患者的风险模型构建 (Life Sci, 2020, 243, 117289)。利用加权基因共表达网络(WGCNA)方法,在系统水平上研究甲状腺癌发病机制中的基因网络特征,将临床特征与基因网络模块结合筛选得到6个病理分期标记基因,图18所示,以此构建晚期患者预后预测模型(Xu Li, Li ML, Guo YZ* et al. Identification of pathological stage-related genes of papillary thyroid cancer along with survival prediction, 审稿中)。 
(4)利用深度学习算法开展基于生物序列及医学图像数据的疾病诊断模型构建。 我们开发了基于卷积神经网络与长短时双向记忆相结合的深度学习模型,对白血病四个亚型TF的7万多个DNA结合位点进行预测 (Chemometr Intell Lab, 2020, 199, 103976 )。轻度认知功能障碍(MCI)作为阿尔茨海默病(AD)的预兆,具有相对复杂多变的特点。针对早、晚期MCI的诊断问题,利用多模态影像学数据核磁共振图像(MRI)和扩散张量图像(DTI)特征,融合临床诊断特征数据,提出了基于3D卷积神经网络的诊断模型,其预测准确率达97.2%,AUC为0.995(Wang C, Pu XM, Guo YZ* et al. A new deep convolutional neural network model for effective prediction of mild cognitive impairment,审稿中)。 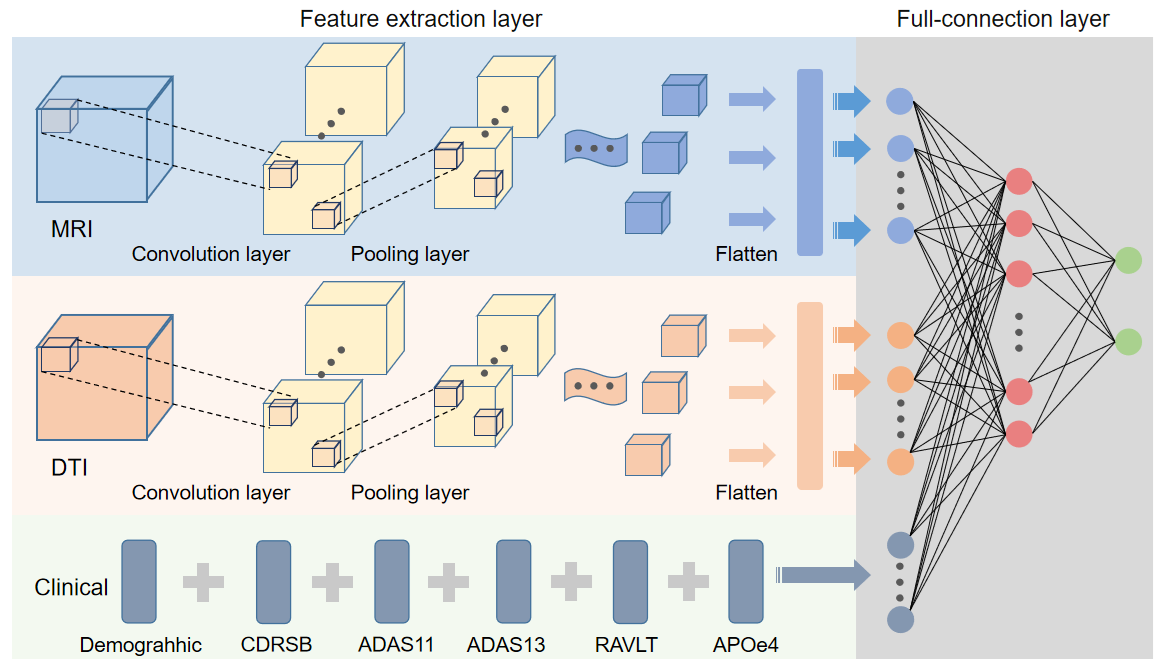
获奖: 2015年教育部自然科学奖二等奖1 项(“典型蛋白质结构功能信息的数字化表征”,排名第二)。 附发表论文列表: 1. Liu Qin,Yanhong Liu, Menglong Li*, Xuemei Pu, Yanzhi Guo*, Briefings in Bioinformatics, 2020, 1, 73-84. 2. Lei Xu, Jian He, Qihang Cai, Xuemei Pu, Yanzhi Guo*, Life Sciences, 2020, 243: 117289. 3. Lei Xu, Lei Zhang, Tian Wang, Yanling Wu, Xuemei Pu, Menglong Li, Yanzhi Guo*, Life Sciences, 2020, 257: 118092. 4. Jian He, Chuan Li*, Xuemei Pu, Menglong Li, Yanzhi Guo*, Chemometrics and Intelligent Laboratory Systems, 2020,199: 103976. 5. Hao Qiu, Yanzhi Guo*, Lezheng Yu, Xuemei Pu, Menglong Li, Chemometrics and Intelligent Laboratory Systems, 2018,179,31-38 6. Wen Hu, Liu Qin, Menglong Li, Xuemei Pu, Yanzhi Guo*,Journal of Computer-Aided Molecular Design,2018,32:1363-1373. 7. W Hu, L Qin, ML Li*, XM Pu, YZ Guo*, RSC Advances, 2018, 8, 10582-10592 8. Yu Wang, Yanzhi Guo*, Xuemei Pu, Menglong Li*, RSC Advances, 2017, 31, 2046-2069. 9. Yu Wang, Yun Lin, Yanzhi Guo*, Xuemei Pu, Menglong Li*, Scientific Reports, 2017,7, 3159 10. Yu Wang, Yanzhi Guo*, Xuemei Pu, Menglong Li, Journal of Computer-Aided Molecular Design,2017, 11, 1029-1038. 11. WL Li, YZ Guo*, ML Li, XM Pu, Interdiscip Sci : Comput life Sci, 2017, 9:459–467 12. Zhongyu Liu, Yanzhi Guo*, Xuemei Pu, Menglong Li*. Scientific Reports, 2016, 6, 34172. 13. Jiesi Luo, Wenling Li, Zhongyu Liu, Yanzhi Guo*, Xuemei Pu, Menglong Li*, Analyst, 2015, 2015,140, 3048-3056. 14. Yayun Hu, Yanzhi Guo*, Yinan Shi, Menglong Li and Xuemei Pu*, RSC Advances, 2015, 5, 42009-42019. 15. Yinanshi, Yanzhi Guo*, Yayun Hu, Menglong Li*, Scientific Reports, 2015, 2015,5,12403. 16. Xu Dai, Runyu Jing, Yanzhi Guo*, Yongcheng Dong, Yuelong Wang, Yuan Liu, Xuemei Pu, Menglong Li*, Current Pharmaceutical Design, 2015, 21, 3051-3061. 17. Jiesi Luo, Zhongyu Liu, Yanzhi Guo*, Menglong Li*, Scientific Reports, 2015,5,14214 18. Yu Wang, Yanzhi Guo*, Qifan Kuang, Xuemei Pu, Yue Ji, Zhihang Zhang, Menglong Li*, Journal of Computer Aided Molecule Design, 2015, 29, 349-360. 19. Yuanyuan Fu, Yanzhi Guo*, Yuelong Wang, Jiesi Luo, Xuemei Pu, Menglong Li* Zhihang Zhang, Computational Biology and Chemistry, 2015, 56, 41-48. 20. Jiesi Luo, Yanzhi Guo*, Yuanyuan Fu, Yu Wang, Wenling Li, Menglong Li*, Proteins: structure, function, and bioinformatics, 2014, 82, 3090–3100. 21. Jiesi Luo, Yanzhi Guo*, Yun Zhong, Duo Ma, Wenling Li, Menglong Li*. Journal of Computer Aided Molecule Design, 2014, 28, 619-629. 22. Duo Ma, Yanzhi Guo*, Jiesi Luo, Xuemei Pu, Menglong Li*, Chemometrics and Intelligent Laboratory Systems, 2014,138, 7-13. 23. Yun Zhong, Yanzhi Guo*, Jiesi Luo, Xuemei Pu, Menglong Li*, Chemometrics and Intelligent Laboratory Systems, 2014,136, 97-103. 24. Xiaojiao Yang, Yanzhi Guo*, Jiesi Luo, Xuemei Pu, Menglong Li*, Plos ONE, 2013, 8, e84439. 25. Wen Liu, Yanzhi Guo*, Jiesi Luo, Yun Zhong, Xiaojiao Yang, Xuemei Pu, Menglong Li*, Chemometrics and Intelligent Laboratory Systems, 2013, 126, 117-122. 26. Lezheng Yu, Jiesi Luo, Yanzhi Guo*, YizhouLi XuemeiPu, Menglong Li*, Computational Biology and Medicine, 2013, 43, 1177-1181. 27. Jiesi Luo, Lezheng Yu, Yanzhi Guo*, Menglong Li*, Chemometr Intellt Lab, 2012,110, 163-167. 28. Wenli Qin, Yizhou Li, Juan Li, Lezheng Yu, Di Wu, Runyu Jing, Xuemei Pu, Yanzh Guo*, Menglong Li*, Computational Biology and Chemistry, 2012, 36, 31-35. 29. Xia Wang, Gang Mi, Cuicui Wang, Yongqing Zhang, Juan Li, Yanzhi Guo*, Xuemei Pu, Menglong Li*, Computational Biology and Medicine, 2012, 42, 1053-1059. 30. Juan Li, Yongqing Zhang, Wenli Qin, Yanzhi Guo*, Lezheng Yu, Xuemei Pu, Menglong Li, Jing Sun. Natural Science, 2012, 4, 304-312. 31. Yanzhi Guo, Menglong Li*, Xuemei Pu, Gongbin Li, Juan Li. BMC Research Notes,2010, 3, 145. 32. Yanzhi Guo, Lezheng Yu, Zhining Wen, Menglong Li. Nucleic Acids Research, 2008, 36, 3025-3030. 33. Yanzhi Guo, Menglong Li, Minchun Lu, Zhining Wen, Kelong Wang, Gongbin Li, Jiang Wu, Amino Acids, 2006, 30, 397-402. 34. Yanzhi Guo, Menglong Li, Minchun Lu, Zhining Wen, Zhongtian Huang. Proteins: structure, function, and bioinformatics, 2006, 65, 55-60. 35. Yanzhi Guo, Menglong Li, Kelong Wang, Zhining Wen, Minchun Lu, Lixia Liu, Lin Jiang. Acta Biochimica et Biophysica Sinica, 2005, 37, 759–766. |
